Ideal for Yeast Cells Homogenization
Do you spend lots of time and effort homogenizing yeast samples? The Bullet Blender® tissue homogenizer delivers high quality and superior yields. No other homogenizer comes close to delivering the Bullet Blender’s winning combination of top-quality performance and budget-friendly affordability. See below for a yeast cells homogenization protocol.
Save Time, Effort and Get Superior Results with
The Bullet Blender Homogenizer
Consistent and High Yield Results
Run up to 24 samples at the same time under microprocessor-controlled conditions, ensuring experimental reproducibility and high yield. Process samples from 10mg or less up to 3.5g.No Cross Contamination
No part of the Bullet Blender ever touches the tissue – the sample tubes are kept closed during homogenization. There are no probes to clean between samples.Samples Stay Cool
The Bullet Blenders’ innovative and elegant design provides convective cooling of the samples, so they do not heat up more than several degrees. In fact, our Gold+ models hold the sample temperature to about 4ºC.Easy and Convenient to Use
Just place beads and buffer along with your tissue sample in standard tubes, load tubes directly in the Bullet Blender, select time and speed, and press start.Risk Free Purchase
Thousands of peer-reviewed journal articles attest to the consistency and quality of the Bullet Blender homogenizer. We offer a 2 year warranty, extendable to 4 years, because our Bullet Blenders are reliable and last for many years.Yeast Cells Homogenization Protocol
Sample size |
See the Protocol |
| microcentrifuge tube model (up to 300 mg) | Small yeast samples |
| 5mL tube model (100mg - 1g) | Medium yeast samples |
| 50mL tube model (100mg - 3.5g) | Large yeast samples |
What Else Can You Homogenize? Tough or Soft, No Problem!
The Bullet Blender can process a wide range of samples including organ tissue, cell culture, plant tissue, and small organisms. You can homogenize samples as tough as mouse femur or for gentle applications such as tissue dissociation or organelle isolation.

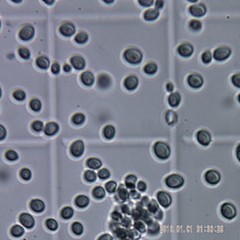
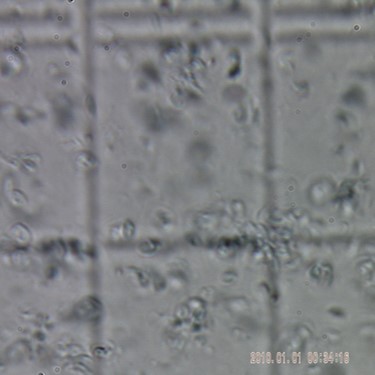
Yeast cells intact (in upper photo) are completely homogenized (in lower photo).
Want more guidance? Need a quote? Contact us:

Bullet Blender Models
Select Publications using the Bullet Blender to Homogenize Yeast Cells
2474232
fungi
1
apa
50
date
desc
3303
https://www.nextadvance.com/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22GQ63XEV2%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Rasool%20et%20al.%22%2C%22parsedDate%22%3A%222016-10-02%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ERasool%2C%20A.%2C%20Ahmed%2C%20M.%20S.%2C%20%26amp%3B%20Li%2C%20C.%20%282016%29.%20Overproduction%20of%20squalene%20synergistically%20downregulates%20ethanol%20production%20in%20Saccharomyces%20cerevisiae.%20%3Ci%3EChemical%20Engineering%20Science%3C%5C%2Fi%3E%2C%20%3Ci%3E152%3C%5C%2Fi%3E%2C%20370%26%23x2013%3B380.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ces.2016.06.014%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ces.2016.06.014%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Overproduction%20of%20squalene%20synergistically%20downregulates%20ethanol%20production%20in%20Saccharomyces%20cerevisiae%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aamir%22%2C%22lastName%22%3A%22Rasool%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Muhammad%20Saad%22%2C%22lastName%22%3A%22Ahmed%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chun%22%2C%22lastName%22%3A%22Li%22%7D%5D%2C%22abstractNote%22%3A%22Metabolic%20engineering%20strategies%20are%20often%20devised%20to%20redirect%20precursor%20flux%20in%20an%20engineered%20pathway.%20To%20develop%20a%20new%20strategy%2C%20the%20effect%20of%20overexpression%20of%20the%20squalene%20biosynthesis%20%28SB%29%20pathway%20and%20squalene%20overproduction%20on%20the%20ethanol%20production%20%28EP%29%20and%20post-squalene%20biosynthesis%20%28PB%29%20pathways%20was%20determined%20in%20Saccharomyces%20cerevisiae.%20Through%20overexpression%20of%20the%20HMG1%2C%20IDI1%2C%20ERG20%20and%20ERG9%20genes%20of%20the%20SB%20pathway%2C%20production%20of%20squalene%20increased%2010-fold%20in%20the%20M1EG%20strain%20compared%20to%20the%20wild-type%20strain%20%28WT%29%20%5B%2834%20mg%5C%2FL%29%2C%20without%20terbinafine%2C%20an%20inhibitor%20of%20squalene%20monooxygenase%2C%20and%2035.02-fold%20%28119.08%20mg%5C%2FL%29%20with%20terbinafine%5D.%20However%2C%20due%20to%20overexpression%20of%20the%20SB%20pathway%20and%20squalene%20overproduction%2C%20production%20of%20ethanol%20and%20functionality%20of%20the%20EP%20and%20PB%20pathways%20were%20synergistically%20downregulated%20by%2051.61%25%2C%2095.86%25%20and%2081.79%25%2C%20respectively%2C%20in%20the%20M1EG%20strain%20compared%20to%20the%20WT%20strain.%20Overexpression%20of%20the%20entire%20SB%20pathway%20also%20enhanced%20production%20of%20squalene%20by%2076.12-fold%20%28304.49%20mg%5C%2FL%29%20and%20synergistically%20downregulated%20production%20of%20ethanol%20and%20functionality%20of%20the%20EP%20and%20PB%20pathways%20by%2066.13%25%2C%2097.02%25%20and%2087.56%25%20in%20the%20FOH-2%20strain%20compared%20to%20the%20WT%20strain%2C%20respectively.%20The%20EP%20and%20PB%20pathways%20were%20strongly%20downregulated%20in%20the%20FOH-2%20strain%20compared%20to%20the%20M1EG%20strain%20because%20the%20FOH-2%20strain%20overexpresses%20the%20entire%20SB%20pathway%20and%20produces%20more%20squalene%20than%20the%20M1EG%20strain.%5Cn%5CnThese%20data%20suggest%20that%20overexpression%20of%20the%20SB%20pathway%20and%20squalene%20overproduction%20downregulate%20the%20EP%20and%20PB%20pathways%20in%20engineered%20strains.%20Therefore%2C%20we%20speculate%20that%20a%20cryptic%20regulation%20mechanism%20may%20downregulate%20these%20pathways%2C%20and%20characterization%20of%20such%20a%20mechanism%20may%20enable%20us%20to%20divert%20the%20precursor%20flux%20from%20the%20EP%20and%20PB%20pathways%20to%20the%20squalene%20biosynthesis%20pathway.%22%2C%22date%22%3A%22October%202%2C%202016%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.ces.2016.06.014%22%2C%22ISSN%22%3A%220009-2509%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.sciencedirect.com%5C%2Fscience%5C%2Farticle%5C%2Fpii%5C%2FS0009250916303062%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-06-24T16%3A36%3A38Z%22%7D%7D%2C%7B%22key%22%3A%22J26QJTXI%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Orgil%20et%20al.%22%2C%22parsedDate%22%3A%222016-06-15%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EOrgil%2C%20O.%2C%20Mor%2C%20H.%2C%20Matityahu%2C%20A.%2C%20%26amp%3B%20Onn%2C%20I.%20%282016%29.%20Identification%20of%20a%20region%20in%20the%20coiled-coil%20domain%20of%20Smc3%20that%20is%20essential%20for%20cohesin%20activity.%20%3Ci%3ENucleic%20Acids%20Research%3C%5C%2Fi%3E%2C%20gkw539.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkw539%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkw539%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Identification%20of%20a%20region%20in%20the%20coiled-coil%20domain%20of%20Smc3%20that%20is%20essential%20for%20cohesin%20activity%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ola%22%2C%22lastName%22%3A%22Orgil%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hadar%22%2C%22lastName%22%3A%22Mor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Avi%22%2C%22lastName%22%3A%22Matityahu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Itay%22%2C%22lastName%22%3A%22Onn%22%7D%5D%2C%22abstractNote%22%3A%22The%20cohesin%20complex%20plays%20an%20important%20role%20in%20sister%20chromatin%20cohesion.%20Cohesin%27s%20core%20is%20composed%20of%20two%20structural%20maintenance%20of%20chromosome%20%28SMC%29%20proteins%2C%20called%20Smc1%20and%20Smc3.%20SMC%20proteins%20are%20built%20from%20a%20globular%20hinge%20domain%2C%20a%20rod-shaped%20domain%20composed%20of%20long%20anti-parallel%20coiled-coil%20%28CC%29%2C%20and%20a%20second%20globular%20adenosine%20triphosphatase%20domain%20called%20the%20head.%20The%20functions%20of%20both%20head%20and%20hinge%20domains%20have%20been%20studied%20extensively%2C%20yet%20the%20function%20of%20the%20CC%20region%20remains%20elusive.%20We%20identified%20a%20mutation%20in%20the%20CC%20of%20smc3%20%28L217P%29%20that%20disrupts%20the%20function%20of%20the%20protein.%20Cells%20carrying%20the%20smc3-L217P%20allele%20have%20a%20strong%20cohesion%20defect%20and%20complexes%20containing%20smc3-L217P%20are%20not%20loaded%20onto%20the%20chromosomes.%20However%2C%20the%20mutation%20does%20not%20affect%20inter-protein%20interactions%20in%20either%20the%20core%20complex%20or%20with%20the%20Scc2%20loader.%20We%20show%20by%20molecular%20dynamics%20and%20biochemistry%20that%20wild-type%20Smc3%20can%20adopt%20distinct%20conformations%2C%20and%20that%20adenosine%20triphosphate%20%28ATP%29%20induces%20the%20conformational%20change.%20The%20L217P%20mutation%20restricts%20the%20ability%20of%20the%20mutated%20protein%20to%20switch%20between%20the%20conformations.%20We%20suggest%20that%20the%20function%20of%20the%20CC%20is%20to%20transfer%20ATP%20binding%5C%2Fhydrolysis%20signals%20between%20the%20head%20and%20the%20hinge%20domains.%20The%20results%20provide%20a%20new%20insight%20into%20the%20mechanism%20of%20cohesin%20activity.%22%2C%22date%22%3A%222016-06-15%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkw539%22%2C%22ISSN%22%3A%220305-1048%2C%201362-4962%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fnar.oxfordjournals.org%5C%2Fcontent%5C%2Fearly%5C%2F2016%5C%2F06%5C%2F15%5C%2Fnar.gkw539%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-06-24T15%3A22%3A15Z%22%7D%7D%2C%7B%22key%22%3A%22VCUJTQ4J%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Shwartz%20et%20al.%22%2C%22parsedDate%22%3A%222016-06-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EShwartz%2C%20M.%2C%20Matityahu%2C%20A.%2C%20%26amp%3B%20Onn%2C%20I.%20%282016%29.%20Identification%20of%20Functional%20Domains%20in%20the%20Cohesin%20Loader%20Subunit%20Scc4%20by%20a%20Random%20Insertion%5C%2FDominant%20Negative%20Screen.%20%3Ci%3EG3%3A%20Genes%7CGenomes%7CGenetics%3C%5C%2Fi%3E%2C%20g3.116.031674.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1534%5C%2Fg3.116.031674%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1534%5C%2Fg3.116.031674%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Identification%20of%20Functional%20Domains%20in%20the%20Cohesin%20Loader%20Subunit%20Scc4%20by%20a%20Random%20Insertion%5C%2FDominant%20Negative%20Screen%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michal%22%2C%22lastName%22%3A%22Shwartz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Avi%22%2C%22lastName%22%3A%22Matityahu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Itay%22%2C%22lastName%22%3A%22Onn%22%7D%5D%2C%22abstractNote%22%3A%22Cohesin%20is%20a%20multi-subunit%20complex%20that%20plays%20an%20essential%20role%20in%20genome%20stability.%20Initial%20association%20of%20cohesin%20with%20chromosomes%20requires%20the%20loader%2C%20a%20heterodimer%20composed%20of%20Scc4%20and%20Scc2.%20However%2C%20very%20little%20is%20known%20about%20the%20loader%27s%20mechanism%20of%20action.%20In%20this%20study%20we%20performed%20a%20genetic%20screen%20to%20identify%20functional%20domains%20in%20the%20Scc4%20subunit%20of%20the%20loader.%20We%20isolated%20scc4%20mutant%20alleles%20that%20when%20overexpressed%20have%20a%20dominant%20negative%20effect%20on%20cell%20viability.%20We%20defined%20a%20small%20region%20in%20the%20N%27%20terminal%20of%20Scc4%20that%20is%20dominant%20negative%20when%20overexpressed%2C%20and%20on%20which%20Scc2%5C%2F4%20activity%20depends.%20When%20the%20mutant%20alleles%20are%20expressed%20as%20a%20single%20copy%2C%20they%20are%20recessive%20and%20do%20not%20support%20cell%20viability%2C%20cohesion%2C%20cohesin%20loading%20or%20Scc4%20chromatin%20binding.%20In%20addition%2C%20we%20show%20that%20the%20mutants%20investigated%20reduce%20but%20do%20not%20eliminate%20the%20interaction%20of%20Scc4%20with%20either%20Scc2%20or%20cohesin.%20However%2C%20we%20show%20that%20Scc4%20cannot%20bind%20cohesin%20in%20the%20absence%20of%20Scc2.%20Our%20results%20provide%20new%20insight%20into%20the%20roles%20of%20Scc4%20in%20cohesin%20loading%20and%20contribute%20to%20the%20deciphering%20of%20the%20loading%20mechanism.%22%2C%22date%22%3A%222016-06-08%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1534%5C%2Fg3.116.031674%22%2C%22ISSN%22%3A%22%2C%202160-1836%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.g3journal.org%5C%2Fcontent%5C%2Fearly%5C%2F2016%5C%2F06%5C%2F06%5C%2Fg3.116.031674%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-06-24T16%3A10%3A35Z%22%7D%7D%2C%7B%22key%22%3A%222MADWFHW%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Zheng%20et%20al.%22%2C%22parsedDate%22%3A%222016-06%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EZheng%2C%20Y.%2C%20Xie%2C%20J.%2C%20Huang%2C%20X.%2C%20Dong%2C%20J.%2C%20Park%2C%20M.%20S.%2C%20%26amp%3B%20Chan%2C%20W.%20K.%20%282016%29.%20Binding%20studies%20using%20Pichia%20pastoris%20expressed%20human%20aryl%20hydrocarbon%20receptor%20and%20aryl%20hydrocarbon%20receptor%20nuclear%20translocator%20proteins.%20%3Ci%3EProtein%20Expression%20and%20Purification%3C%5C%2Fi%3E%2C%20%3Ci%3E122%3C%5C%2Fi%3E%2C%2072%26%23x2013%3B81.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.pep.2016.02.011%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.pep.2016.02.011%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Binding%20studies%20using%20Pichia%20pastoris%20expressed%20human%20aryl%20hydrocarbon%20receptor%20and%20aryl%20hydrocarbon%20receptor%20nuclear%20translocator%20proteins%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yujuan%22%2C%22lastName%22%3A%22Zheng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jinghang%22%2C%22lastName%22%3A%22Xie%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xin%22%2C%22lastName%22%3A%22Huang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jin%22%2C%22lastName%22%3A%22Dong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Miki%20S.%22%2C%22lastName%22%3A%22Park%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22William%20K.%22%2C%22lastName%22%3A%22Chan%22%7D%5D%2C%22abstractNote%22%3A%22The%20aryl%20hydrocarbon%20receptor%20%28AHR%29%20is%20a%20transcription%20factor%20which%20activates%20gene%20transcription%20by%20binding%20to%20its%20corresponding%20enhancer%20as%20the%20heterodimer%2C%20which%20is%20consisted%20of%20AHR%20and%20the%20aryl%20hydrocarbon%20receptor%20nuclear%20translocator%20%28ARNT%29.%20Human%20AHR%20can%20be%20rather%20difficult%20to%20study%2C%20when%20compared%20among%20the%20AHR%20of%20other%20species%2C%20since%20it%20is%20relatively%20unstable%20and%20less%20sensitive%20to%20some%20ligands%20in%5Cu00a0vitro.%20Overexpression%20of%20human%20AHR%20has%20been%20limited%20to%20the%20baculovirus%20expression%2C%20which%20is%20costly%20and%20tedious%20due%20to%20the%20need%20of%20repetitive%20baculovirus%20production.%20Here%20we%20explored%20whether%20we%20could%20generate%20abundant%20amounts%20of%20human%20AHR%20and%20ARNT%20in%20a%20better%20overexpression%20system%20for%20functional%20study.%20We%20observed%20that%20human%20AHR%20and%20ARNT%20can%20be%20expressed%20in%20Pichia%20pastoris%20with%20yields%20that%20are%20comparable%20to%20the%20baculovirus%20system%20only%20if%20their%20cDNAs%20are%20optimized%20for%20Pichia%20expression.%20Fusion%20with%20a%20c-myc%20tag%20at%20their%20C-termini%20seems%20to%20increase%20the%20expression%20yield.%20These%20Pichia%20expressed%20proteins%20can%20effectively%20heterodimerize%20and%20form%20the%20ternary%20AHR%5C%2FARNT%5C%2Fenhancer%20complex%20in%20the%20presence%20of%20%5Cu03b2-naphthoflavone%20or%20kynurenine.%20Limited%20proteolysis%20using%20thermolysin%20can%20be%20used%20to%20study%20the%20heterodimerization%20of%20these%20human%20AHR%20and%20ARNT%20proteins.%22%2C%22date%22%3A%22June%202016%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.pep.2016.02.011%22%2C%22ISSN%22%3A%221046-5928%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.sciencedirect.com%5C%2Fscience%5C%2Farticle%5C%2Fpii%5C%2FS1046592816300316%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-06-24T15%3A24%3A33Z%22%7D%7D%2C%7B%22key%22%3A%224DI9RTGZ%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Mashock%22%2C%22parsedDate%22%3A%222016-05%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMashock%2C%20M.%20J.%20%282016%29.%20%3Ci%3EExamining%20mechanism%20of%20toxicity%20of%20copper%20oxide%20nanoparticles%20to%20Saccharomyces%20cerevisiae%20and%20Caenorhabditis%20elegans%3C%5C%2Fi%3E.%20Marquette%20University.%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22thesis%22%2C%22title%22%3A%22Examining%20mechanism%20of%20toxicity%20of%20copper%20oxide%20nanoparticles%20to%20Saccharomyces%20cerevisiae%20and%20Caenorhabditis%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20Joseph%22%2C%22lastName%22%3A%22Mashock%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22thesisType%22%3A%22%22%2C%22university%22%3A%22Marquette%20University%22%2C%22date%22%3A%22May%202016%22%2C%22language%22%3A%22%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-06-29T19%3A51%3A46Z%22%7D%7D%2C%7B%22key%22%3A%2263E7XRAQ%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Avbelj%20et%20al.%22%2C%22parsedDate%22%3A%222015-09-30%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EAvbelj%2C%20M.%2C%20Zupan%2C%20J.%2C%20Kranjc%2C%20L.%2C%20%26amp%3B%20Raspor%2C%20P.%20%282015%29.%20Quorum-Sensing%20Kinetics%20in%20%3Ci%3ESaccharomyces%20cerevisiae%3C%5C%2Fi%3E%26%23x202F%3B%3A%20A%20Symphony%20of%20%3Ci%3EARO%3C%5C%2Fi%3E%20Genes%20and%20Aromatic%20Alcohols.%20%3Ci%3EJournal%20of%20Agricultural%20and%20Food%20Chemistry%3C%5C%2Fi%3E%2C%20%3Ci%3E63%3C%5C%2Fi%3E%2838%29%2C%208544%26%23x2013%3B8550.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facs.jafc.5b03400%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facs.jafc.5b03400%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Quorum-Sensing%20Kinetics%20in%20%3Ci%3ESaccharomyces%20cerevisiae%3C%5C%2Fi%3E%20%3A%20A%20Symphony%20of%20%3Ci%3EARO%3C%5C%2Fi%3E%20Genes%20and%20Aromatic%20Alcohols%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Martina%22%2C%22lastName%22%3A%22Avbelj%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jure%22%2C%22lastName%22%3A%22Zupan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Luka%22%2C%22lastName%22%3A%22Kranjc%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Raspor%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-09-30%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1021%5C%2Facs.jafc.5b03400%22%2C%22ISSN%22%3A%220021-8561%2C%201520-5118%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fpubs.acs.org%5C%2Fdoi%5C%2F10.1021%5C%2Facs.jafc.5b03400%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-10-30T20%3A17%3A01Z%22%7D%7D%2C%7B%22key%22%3A%22GTXA48W3%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Meyer%20et%20al.%22%2C%22parsedDate%22%3A%222015-09-01%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMeyer%2C%20R.%20E.%2C%20Chuong%2C%20H.%20H.%2C%20Hild%2C%20M.%2C%20Hansen%2C%20C.%20L.%2C%20Kinter%2C%20M.%2C%20%26amp%3B%20Dawson%2C%20D.%20S.%20%282015%29.%20Ipl1%5C%2FAurora-B%20is%20necessary%20for%20kinetochore%20restructuring%20in%20meiosis%20I%20in%20Saccharomyces%20cerevisiae.%20%3Ci%3EMolecular%20Biology%20of%20the%20Cell%3C%5C%2Fi%3E%2C%20%3Ci%3E26%3C%5C%2Fi%3E%2817%29%2C%202986%26%23x2013%3B3000.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1091%5C%2Fmbc.E15-01-0032%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1091%5C%2Fmbc.E15-01-0032%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Ipl1%5C%2FAurora-B%20is%20necessary%20for%20kinetochore%20restructuring%20in%20meiosis%20I%20in%20Saccharomyces%20cerevisiae%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20E.%22%2C%22lastName%22%3A%22Meyer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22H.%20H.%22%2C%22lastName%22%3A%22Chuong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Hild%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%20L.%22%2C%22lastName%22%3A%22Hansen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Kinter%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20S.%22%2C%22lastName%22%3A%22Dawson%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-09-01%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1091%5C%2Fmbc.E15-01-0032%22%2C%22ISSN%22%3A%221059-1524%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.molbiolcell.org%5C%2Fcgi%5C%2Fdoi%5C%2F10.1091%5C%2Fmbc.E15-01-0032%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-12-31T21%3A13%3A17Z%22%7D%7D%2C%7B%22key%22%3A%226XP87W28%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Aylward%20et%20al.%22%2C%22parsedDate%22%3A%222015-08-28%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EAylward%2C%20F.%20O.%2C%20Khadempour%2C%20L.%2C%20Tremmel%2C%20D.%20M.%2C%20McDonald%2C%20B.%20R.%2C%20Nicora%2C%20C.%20D.%2C%20Wu%2C%20S.%2C%20Moore%2C%20R.%20J.%2C%20Orton%2C%20D.%20J.%2C%20Monroe%2C%20M.%20E.%2C%20Piehowski%2C%20P.%20D.%2C%20Purvine%2C%20S.%20O.%2C%20Smith%2C%20R.%20D.%2C%20Lipton%2C%20M.%20S.%2C%20Burnum-Johnson%2C%20K.%20E.%2C%20%26amp%3B%20Currie%2C%20C.%20R.%20%282015%29.%20Enrichment%20and%20Broad%20Representation%20of%20Plant%20Biomass-Degrading%20Enzymes%20in%20the%20Specialized%20Hyphal%20Swellings%20of%20Leucoagaricus%20gongylophorus%2C%20the%20Fungal%20Symbiont%20of%20Leaf-Cutter%20Ants.%20%3Ci%3EPLOS%20ONE%3C%5C%2Fi%3E%2C%20%3Ci%3E10%3C%5C%2Fi%3E%288%29%2C%20e0134752.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pone.0134752%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pone.0134752%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Enrichment%20and%20Broad%20Representation%20of%20Plant%20Biomass-Degrading%20Enzymes%20in%20the%20Specialized%20Hyphal%20Swellings%20of%20Leucoagaricus%20gongylophorus%2C%20the%20Fungal%20Symbiont%20of%20Leaf-Cutter%20Ants%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Frank%20O.%22%2C%22lastName%22%3A%22Aylward%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lily%22%2C%22lastName%22%3A%22Khadempour%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%20M.%22%2C%22lastName%22%3A%22Tremmel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bradon%20R.%22%2C%22lastName%22%3A%22McDonald%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Carrie%20D.%22%2C%22lastName%22%3A%22Nicora%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Si%22%2C%22lastName%22%3A%22Wu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ronald%20J.%22%2C%22lastName%22%3A%22Moore%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%20J.%22%2C%22lastName%22%3A%22Orton%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Matthew%20E.%22%2C%22lastName%22%3A%22Monroe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%20D.%22%2C%22lastName%22%3A%22Piehowski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Samuel%20O.%22%2C%22lastName%22%3A%22Purvine%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Richard%20D.%22%2C%22lastName%22%3A%22Smith%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mary%20S.%22%2C%22lastName%22%3A%22Lipton%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kristin%20E.%22%2C%22lastName%22%3A%22Burnum-Johnson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cameron%20R.%22%2C%22lastName%22%3A%22Currie%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Sean%22%2C%22lastName%22%3A%22Brady%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-8-28%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1371%5C%2Fjournal.pone.0134752%22%2C%22ISSN%22%3A%221932-6203%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fdx.plos.org%5C%2F10.1371%5C%2Fjournal.pone.0134752%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-10-29T21%3A45%3A36Z%22%7D%7D%2C%7B%22key%22%3A%22QA9T8G2K%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Desai%20et%20al.%22%2C%22parsedDate%22%3A%222015-07-24%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EDesai%2C%20J.%2C%20Cheng%2C%20S.%2C%20Ying%2C%20T.%2C%20Nguyen%2C%20M.%2C%20Clancy%2C%20C.%2C%20Lanni%2C%20F.%2C%20%26amp%3B%20Mitchell%2C%20A.%20%282015%29.%20Coordination%20of%20Candida%20albicans%20Invasion%20and%20Infection%20Functions%20by%20Phosphoglycerol%20Phosphatase%20Rhr2.%20%3Ci%3EPathogens%3C%5C%2Fi%3E%2C%20%3Ci%3E4%3C%5C%2Fi%3E%283%29%2C%20573%26%23x2013%3B589.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fpathogens4030573%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fpathogens4030573%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Coordination%20of%20Candida%20albicans%20Invasion%20and%20Infection%20Functions%20by%20Phosphoglycerol%20Phosphatase%20Rhr2%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jigar%22%2C%22lastName%22%3A%22Desai%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shaoji%22%2C%22lastName%22%3A%22Cheng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tammy%22%2C%22lastName%22%3A%22Ying%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Nguyen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cornelius%22%2C%22lastName%22%3A%22Clancy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Frederick%22%2C%22lastName%22%3A%22Lanni%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aaron%22%2C%22lastName%22%3A%22Mitchell%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-07-24%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.3390%5C%2Fpathogens4030573%22%2C%22ISSN%22%3A%222076-0817%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.mdpi.com%5C%2F2076-0817%5C%2F4%5C%2F3%5C%2F573%5C%2F%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-12-31T21%3A41%3A55Z%22%7D%7D%2C%7B%22key%22%3A%22RDBF8WQ4%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Zhang%20et%20al.%22%2C%22parsedDate%22%3A%222015-03-31%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EZhang%2C%20L.%2C%20Li%2C%20X.%2C%20Hill%2C%20R.%20C.%2C%20Qiu%2C%20Y.%2C%20Zhang%2C%20W.%2C%20Hansen%2C%20K.%20C.%2C%20%26amp%3B%20Zhao%2C%20R.%20%282015%29.%20Brr2%20plays%20a%20role%20in%20spliceosomal%20activation%20in%20addition%20to%20U4%5C%2FU6%20unwinding.%20%3Ci%3ENucleic%20Acids%20Research%3C%5C%2Fi%3E%2C%20%3Ci%3E43%3C%5C%2Fi%3E%286%29%2C%203286%26%23x2013%3B3297.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkv062%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkv062%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Brr2%20plays%20a%20role%20in%20spliceosomal%20activation%20in%20addition%20to%20U4%5C%2FU6%20unwinding%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%22%2C%22lastName%22%3A%22Zhang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22X.%22%2C%22lastName%22%3A%22Li%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20C.%22%2C%22lastName%22%3A%22Hill%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y.%22%2C%22lastName%22%3A%22Qiu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22W.%22%2C%22lastName%22%3A%22Zhang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22K.%20C.%22%2C%22lastName%22%3A%22Hansen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%22%2C%22lastName%22%3A%22Zhao%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-03-31%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkv062%22%2C%22ISSN%22%3A%220305-1048%2C%201362-4962%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fnar.oxfordjournals.org%5C%2Flookup%5C%2Fdoi%5C%2F10.1093%5C%2Fnar%5C%2Fgkv062%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-12-31T19%3A44%3A28Z%22%7D%7D%2C%7B%22key%22%3A%22AD3QC58U%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Orgil%20et%20al.%22%2C%22parsedDate%22%3A%222015-03-06%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EOrgil%2C%20O.%2C%20Matityahu%2C%20A.%2C%20Eng%2C%20T.%2C%20Guacci%2C%20V.%2C%20Koshland%2C%20D.%2C%20%26amp%3B%20Onn%2C%20I.%20%282015%29.%20A%20Conserved%20Domain%20in%20the%20Scc3%20Subunit%20of%20Cohesin%20Mediates%20the%20Interaction%20with%20Both%20Mcd1%20and%20the%20Cohesin%20Loader%20Complex.%20%3Ci%3EPLOS%20Genetics%3C%5C%2Fi%3E%2C%20%3Ci%3E11%3C%5C%2Fi%3E%283%29%2C%20e1005036.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1005036%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1005036%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20Conserved%20Domain%20in%20the%20Scc3%20Subunit%20of%20Cohesin%20Mediates%20the%20Interaction%20with%20Both%20Mcd1%20and%20the%20Cohesin%20Loader%20Complex%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ola%22%2C%22lastName%22%3A%22Orgil%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Avi%22%2C%22lastName%22%3A%22Matityahu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Eng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Guacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Douglas%22%2C%22lastName%22%3A%22Koshland%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Itay%22%2C%22lastName%22%3A%22Onn%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Rolf%22%2C%22lastName%22%3A%22Jessberger%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222015-3-6%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1371%5C%2Fjournal.pgen.1005036%22%2C%22ISSN%22%3A%221553-7404%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fdx.plos.org%5C%2F10.1371%5C%2Fjournal.pgen.1005036%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222016-01-04T21%3A44%3A28Z%22%7D%7D%2C%7B%22key%22%3A%224GZ5KIBG%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kurtzman%20and%20Robnett%22%2C%22parsedDate%22%3A%222014%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKurtzman%2C%20C.%20P.%2C%20%26amp%3B%20Robnett%2C%20C.%20J.%20%282014%29.%20Three%20new%20anascosporic%20genera%20of%20the%20Saccharomycotina%3A%20Danielozyma%20gen.%20nov.%2C%20Deakozyma%20gen.%20nov.%20and%20Middelhovenomyces%20gen.%20nov.%20%3Ci%3EAntonie%20van%20Leeuwenhoek%3C%5C%2Fi%3E%2C%20%3Ci%3E105%3C%5C%2Fi%3E%285%29%2C%20933%26%23x2013%3B942.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs10482-014-0149-9%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs10482-014-0149-9%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Three%20new%20anascosporic%20genera%20of%20the%20Saccharomycotina%3A%20Danielozyma%20gen.%20nov.%2C%20Deakozyma%20gen.%20nov.%20and%20Middelhovenomyces%20gen.%20nov%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cletus%20P.%22%2C%22lastName%22%3A%22Kurtzman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christie%20J.%22%2C%22lastName%22%3A%22Robnett%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%225%5C%2F2014%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1007%5C%2Fs10482-014-0149-9%22%2C%22ISSN%22%3A%220003-6072%2C%201572-9699%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Flink.springer.com%5C%2F10.1007%5C%2Fs10482-014-0149-9%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-08-03T15%3A37%3A40Z%22%7D%7D%2C%7B%22key%22%3A%22WIJK22KQ%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Wiedner%20et%20al.%22%2C%22parsedDate%22%3A%222013-07-01%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EWiedner%2C%20S.%20D.%2C%20Ansong%2C%20C.%2C%20Webb-Robertson%2C%20B.-J.%2C%20Pederson%2C%20L.%20M.%2C%20Fortuin%2C%20S.%2C%20Hofstad%2C%20B.%20A.%2C%20Shukla%2C%20A.%20K.%2C%20Panisko%2C%20E.%20A.%2C%20Smith%2C%20R.%20D.%2C%20%26amp%3B%20Wright%2C%20A.%20T.%20%282013%29.%20Disparate%20Proteome%20Responses%20of%20Pathogenic%20and%20Nonpathogenic%20Aspergilli%20to%20Human%20Serum%20Measured%20by%20Activity-Based%20Protein%20Profiling%20%28ABPP%29.%20%3Ci%3EMolecular%20%26amp%3B%20Cellular%20Proteomics%3C%5C%2Fi%3E%2C%20%3Ci%3E12%3C%5C%2Fi%3E%287%29%2C%201791%26%23x2013%3B1805.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fmcp.M112.026534%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fmcp.M112.026534%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Disparate%20Proteome%20Responses%20of%20Pathogenic%20and%20Nonpathogenic%20Aspergilli%20to%20Human%20Serum%20Measured%20by%20Activity-Based%20Protein%20Profiling%20%28ABPP%29%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20D.%22%2C%22lastName%22%3A%22Wiedner%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%22%2C%22lastName%22%3A%22Ansong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B.-J.%22%2C%22lastName%22%3A%22Webb-Robertson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20M.%22%2C%22lastName%22%3A%22Pederson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Fortuin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B.%20A.%22%2C%22lastName%22%3A%22Hofstad%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%20K.%22%2C%22lastName%22%3A%22Shukla%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22E.%20A.%22%2C%22lastName%22%3A%22Panisko%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20D.%22%2C%22lastName%22%3A%22Smith%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%20T.%22%2C%22lastName%22%3A%22Wright%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222013-07-01%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1074%5C%2Fmcp.M112.026534%22%2C%22ISSN%22%3A%221535-9476%2C%201535-9484%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.mcponline.org%5C%2Fcgi%5C%2Fdoi%5C%2F10.1074%5C%2Fmcp.M112.026534%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-07-09T15%3A18%3A11Z%22%7D%7D%2C%7B%22key%22%3A%22B7JEKX55%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Wiedner%20et%20al.%22%2C%22parsedDate%22%3A%222012-09-28%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EWiedner%2C%20S.%20D.%2C%20Burnum%2C%20K.%20E.%2C%20Pederson%2C%20L.%20M.%2C%20Anderson%2C%20L.%20N.%2C%20Fortuin%2C%20S.%2C%20Chauvigne-Hines%2C%20L.%20M.%2C%20Shukla%2C%20A.%20K.%2C%20Ansong%2C%20C.%2C%20Panisko%2C%20E.%20A.%2C%20Smith%2C%20R.%20D.%2C%20%26amp%3B%20Wright%2C%20A.%20T.%20%282012%29.%20Multiplexed%20Activity-based%20Protein%20Profiling%20of%20the%20Human%20Pathogen%20Aspergillus%20fumigatus%20Reveals%20Large%20Functional%20Changes%20upon%20Exposure%20to%20Human%20Serum.%20%3Ci%3EJournal%20of%20Biological%20Chemistry%3C%5C%2Fi%3E%2C%20%3Ci%3E287%3C%5C%2Fi%3E%2840%29%2C%2033447%26%23x2013%3B33459.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fjbc.M112.394106%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fjbc.M112.394106%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Multiplexed%20Activity-based%20Protein%20Profiling%20of%20the%20Human%20Pathogen%20Aspergillus%20fumigatus%20Reveals%20Large%20Functional%20Changes%20upon%20Exposure%20to%20Human%20Serum%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20D.%22%2C%22lastName%22%3A%22Wiedner%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22K.%20E.%22%2C%22lastName%22%3A%22Burnum%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20M.%22%2C%22lastName%22%3A%22Pederson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20N.%22%2C%22lastName%22%3A%22Anderson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Fortuin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20M.%22%2C%22lastName%22%3A%22Chauvigne-Hines%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%20K.%22%2C%22lastName%22%3A%22Shukla%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%22%2C%22lastName%22%3A%22Ansong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22E.%20A.%22%2C%22lastName%22%3A%22Panisko%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20D.%22%2C%22lastName%22%3A%22Smith%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%20T.%22%2C%22lastName%22%3A%22Wright%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222012-09-28%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1074%5C%2Fjbc.M112.394106%22%2C%22ISSN%22%3A%220021-9258%2C%201083-351X%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.jbc.org%5C%2Fcgi%5C%2Fdoi%5C%2F10.1074%5C%2Fjbc.M112.394106%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-07-09T14%3A58%3A29Z%22%7D%7D%2C%7B%22key%22%3A%22WVQDAF73%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Yin%20et%20al.%22%2C%22parsedDate%22%3A%222012%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EYin%2C%20Y.%2C%20Liu%2C%20Y.%2C%20Jin%2C%20H.%2C%20Wang%2C%20S.%2C%20Zhao%2C%20S.%2C%20Geng%2C%20X.%2C%20Li%2C%20M.%2C%20%26amp%3B%20Xu%2C%20F.%20%282012%29.%20Polyethylene%20glycol-mediated%20transformation%20of%20fused%20egfp-hph%20gene%20under%20the%20control%20of%20gpd%20promoter%20in%20Pleurotus%20eryngii.%20%3Ci%3EBiotechnology%20Letters%3C%5C%2Fi%3E%2C%20%3Ci%3E34%3C%5C%2Fi%3E%2810%29%2C%201895%26%23x2013%3B1900.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs10529-012-0985-5%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs10529-012-0985-5%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Polyethylene%20glycol-mediated%20transformation%20of%20fused%20egfp-hph%20gene%20under%20the%20control%20of%20gpd%20promoter%20in%20Pleurotus%20eryngii%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yonggang%22%2C%22lastName%22%3A%22Yin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yu%22%2C%22lastName%22%3A%22Liu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Haojie%22%2C%22lastName%22%3A%22Jin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shouxian%22%2C%22lastName%22%3A%22Wang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shuang%22%2C%22lastName%22%3A%22Zhao%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xiaoli%22%2C%22lastName%22%3A%22Geng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ming%22%2C%22lastName%22%3A%22Li%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Feng%22%2C%22lastName%22%3A%22Xu%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%2210%5C%2F2012%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1007%5C%2Fs10529-012-0985-5%22%2C%22ISSN%22%3A%220141-5492%2C%201573-6776%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Flink.springer.com%5C%2F10.1007%5C%2Fs10529-012-0985-5%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-07-09T15%3A14%3A14Z%22%7D%7D%2C%7B%22key%22%3A%22M3GMSWHC%22%2C%22library%22%3A%7B%22id%22%3A2474232%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Suzuki%20et%20al.%22%2C%22parsedDate%22%3A%222012%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ESuzuki%2C%20Y.%2C%20Murray%2C%20S.%20L.%2C%20Wong%2C%20K.%20H.%2C%20Davis%2C%20M.%20A.%2C%20%26amp%3B%20Hynes%2C%20M.%20J.%20%282012%29.%20Reprogramming%20of%20carbon%20metabolism%20by%20the%20transcriptional%20activators%20AcuK%20and%20AcuM%20in%20Aspergillus%20nidulans%3A%20Transcription%20factors%20controlling%20carbon%20metabolism.%20%3Ci%3EMolecular%20Microbiology%3C%5C%2Fi%3E%2C%20%3Ci%3E84%3C%5C%2Fi%3E%285%29%2C%20942%26%23x2013%3B964.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fj.1365-2958.2012.08067.x%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fj.1365-2958.2012.08067.x%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Reprogramming%20of%20carbon%20metabolism%20by%20the%20transcriptional%20activators%20AcuK%20and%20AcuM%20in%20Aspergillus%20nidulans%3A%20Transcription%20factors%20controlling%20carbon%20metabolism%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yumi%22%2C%22lastName%22%3A%22Suzuki%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandra%20L.%22%2C%22lastName%22%3A%22Murray%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Koon%20Ho%22%2C%22lastName%22%3A%22Wong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Meryl%20A.%22%2C%22lastName%22%3A%22Davis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20J.%22%2C%22lastName%22%3A%22Hynes%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%2206%5C%2F2012%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1111%5C%2Fj.1365-2958.2012.08067.x%22%2C%22ISSN%22%3A%220950382X%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fdoi.wiley.com%5C%2F10.1111%5C%2Fj.1365-2958.2012.08067.x%22%2C%22collections%22%3A%5B%22M2MNG549%22%5D%2C%22dateModified%22%3A%222015-07-09T15%3A06%3A47Z%22%7D%7D%5D%7D
Rasool, A., Ahmed, M. S., & Li, C. (2016). Overproduction of squalene synergistically downregulates ethanol production in Saccharomyces cerevisiae. Chemical Engineering Science, 152, 370–380. https://doi.org/10.1016/j.ces.2016.06.014
Orgil, O., Mor, H., Matityahu, A., & Onn, I. (2016). Identification of a region in the coiled-coil domain of Smc3 that is essential for cohesin activity. Nucleic Acids Research, gkw539. https://doi.org/10.1093/nar/gkw539
Shwartz, M., Matityahu, A., & Onn, I. (2016). Identification of Functional Domains in the Cohesin Loader Subunit Scc4 by a Random Insertion/Dominant Negative Screen. G3: Genes|Genomes|Genetics, g3.116.031674. https://doi.org/10.1534/g3.116.031674
Zheng, Y., Xie, J., Huang, X., Dong, J., Park, M. S., & Chan, W. K. (2016). Binding studies using Pichia pastoris expressed human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins. Protein Expression and Purification, 122, 72–81. https://doi.org/10.1016/j.pep.2016.02.011
Mashock, M. J. (2016). Examining mechanism of toxicity of copper oxide nanoparticles to Saccharomyces cerevisiae and Caenorhabditis elegans. Marquette University.
Avbelj, M., Zupan, J., Kranjc, L., & Raspor, P. (2015). Quorum-Sensing Kinetics in Saccharomyces cerevisiae : A Symphony of ARO Genes and Aromatic Alcohols. Journal of Agricultural and Food Chemistry, 63(38), 8544–8550. https://doi.org/10.1021/acs.jafc.5b03400
Meyer, R. E., Chuong, H. H., Hild, M., Hansen, C. L., Kinter, M., & Dawson, D. S. (2015). Ipl1/Aurora-B is necessary for kinetochore restructuring in meiosis I in Saccharomyces cerevisiae. Molecular Biology of the Cell, 26(17), 2986–3000. https://doi.org/10.1091/mbc.E15-01-0032
Aylward, F. O., Khadempour, L., Tremmel, D. M., McDonald, B. R., Nicora, C. D., Wu, S., Moore, R. J., Orton, D. J., Monroe, M. E., Piehowski, P. D., Purvine, S. O., Smith, R. D., Lipton, M. S., Burnum-Johnson, K. E., & Currie, C. R. (2015). Enrichment and Broad Representation of Plant Biomass-Degrading Enzymes in the Specialized Hyphal Swellings of Leucoagaricus gongylophorus, the Fungal Symbiont of Leaf-Cutter Ants. PLOS ONE, 10(8), e0134752. https://doi.org/10.1371/journal.pone.0134752
Desai, J., Cheng, S., Ying, T., Nguyen, M., Clancy, C., Lanni, F., & Mitchell, A. (2015). Coordination of Candida albicans Invasion and Infection Functions by Phosphoglycerol Phosphatase Rhr2. Pathogens, 4(3), 573–589. https://doi.org/10.3390/pathogens4030573
Zhang, L., Li, X., Hill, R. C., Qiu, Y., Zhang, W., Hansen, K. C., & Zhao, R. (2015). Brr2 plays a role in spliceosomal activation in addition to U4/U6 unwinding. Nucleic Acids Research, 43(6), 3286–3297. https://doi.org/10.1093/nar/gkv062
Orgil, O., Matityahu, A., Eng, T., Guacci, V., Koshland, D., & Onn, I. (2015). A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex. PLOS Genetics, 11(3), e1005036. https://doi.org/10.1371/journal.pgen.1005036
Kurtzman, C. P., & Robnett, C. J. (2014). Three new anascosporic genera of the Saccharomycotina: Danielozyma gen. nov., Deakozyma gen. nov. and Middelhovenomyces gen. nov. Antonie van Leeuwenhoek, 105(5), 933–942. https://doi.org/10.1007/s10482-014-0149-9
Wiedner, S. D., Ansong, C., Webb-Robertson, B.-J., Pederson, L. M., Fortuin, S., Hofstad, B. A., Shukla, A. K., Panisko, E. A., Smith, R. D., & Wright, A. T. (2013). Disparate Proteome Responses of Pathogenic and Nonpathogenic Aspergilli to Human Serum Measured by Activity-Based Protein Profiling (ABPP). Molecular & Cellular Proteomics, 12(7), 1791–1805. https://doi.org/10.1074/mcp.M112.026534
Wiedner, S. D., Burnum, K. E., Pederson, L. M., Anderson, L. N., Fortuin, S., Chauvigne-Hines, L. M., Shukla, A. K., Ansong, C., Panisko, E. A., Smith, R. D., & Wright, A. T. (2012). Multiplexed Activity-based Protein Profiling of the Human Pathogen Aspergillus fumigatus Reveals Large Functional Changes upon Exposure to Human Serum. Journal of Biological Chemistry, 287(40), 33447–33459. https://doi.org/10.1074/jbc.M112.394106
Yin, Y., Liu, Y., Jin, H., Wang, S., Zhao, S., Geng, X., Li, M., & Xu, F. (2012). Polyethylene glycol-mediated transformation of fused egfp-hph gene under the control of gpd promoter in Pleurotus eryngii. Biotechnology Letters, 34(10), 1895–1900. https://doi.org/10.1007/s10529-012-0985-5
Suzuki, Y., Murray, S. L., Wong, K. H., Davis, M. A., & Hynes, M. J. (2012). Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans: Transcription factors controlling carbon metabolism. Molecular Microbiology, 84(5), 942–964. https://doi.org/10.1111/j.1365-2958.2012.08067.x






